Ben Cardoen
Computational scientist designing novel weakly supervised reconstruction and detection algorithms in superresolution microscopy.
I design novel weakly supervised interaction analysis algorithms on volumetric and point cloud data from superresolution microscopy.
I leverage my background in parallel and distributed discrete event simulators PDEVS (B.Sc.), meta- and hyperheuristics, and symbolic regression (M. Sc.) in my current research.
My current focus is extending probabilistic learning algorithms to reconstruct the interaction between proteins and organelles, often below the limit of the acquisition, in multichannel superresolution microscopy on scales from 100 to 10nm.
The size of the data necessitates that my algorithms are scalable, leveraging both shared memory parallelism and distributed computing, often on SLURM based supercomputers.
I extend weakly supervised learning using belief theory to support nested labels, recover equilibria transitions, and leverage closed form upper and lower boundaries to the probability of each object supporting a subset of the labels.
Labels can in turn be genomic, relate to viral infection, drug treatment, or environmental stressors to the cell, or a combination of both.
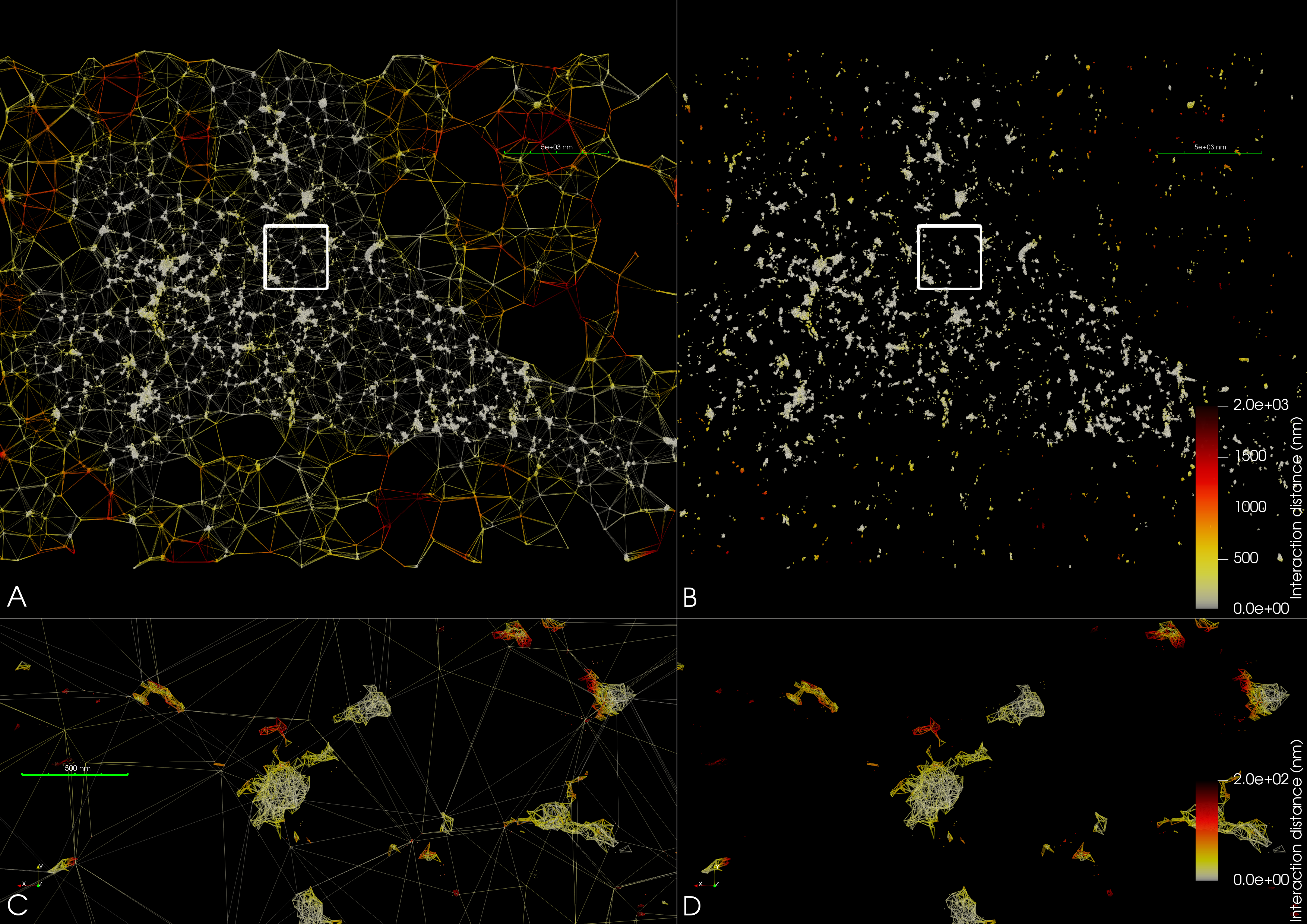
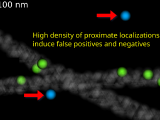
The 3D rendering shows a discrete vectorfield interaction graph between two SMLM point cloud datasets, see publications for the relevant papers.
PDFs of my publications are publicly available , while a more in depth description of the projects is listed here.
I post on both research, philosophy, and technical problems in blog posts.